→ Plot CCF vs Time
This plot shows the cross-correlation functions (CCF) vs time. The parameters
allow to plot the daily or the mov-stacked CCF. Filters and components are
selectable too. The --ampli argument allows to increase the vertical scale
of the CCFs. The --seismic shows the up-going wiggles with a black-filled
background (very heavy !). Passing --refilter allows to bandpass filter
CCFs before plotting (new in 1.5).
msnoise cc plot ccftime --help
Usage: [OPTIONS] STA1 STA2 [EXTRA_ARGS]...
Plots the ccf vs time between sta1 and sta2 STA1 and STA2 must be provided
with this format: NET.STA.LOC !
Options:
-f, --filterid INTEGER Filter ID
-c, --comp TEXT Components (ZZ, ZE, NZ, 1E,...). Defaults to ZZ
-m, --mov_stack INTEGER Mov Stack to read from disk. Defaults to 1.
-a, --ampli FLOAT Amplification of the individual lines on the
vertical axis (default=1)
-S, --seismic Seismic style: fill the space between the zero and
the positive wiggles
-s, --show BOOLEAN Show interactively?
-o, --outfile TEXT Output filename (?=auto). Defaults to PNG format,
but can be anything matplotlib outputs, e.g. ?.pdf
will save to PDF with an automatic file naming.
-e, --envelope Plot envelope instead of time series
-r, --refilter TEXT Refilter CCFs before plotting (e.g. 4:8 for
filtering CCFs between 4.0 and 8.0 Hz. This will
update the plot title.
--normalize TEXT
--help Show this message and exit.
Example:
msnoise cc plot ccftime YA.UV06 YA.UV11 will plot all defaults:
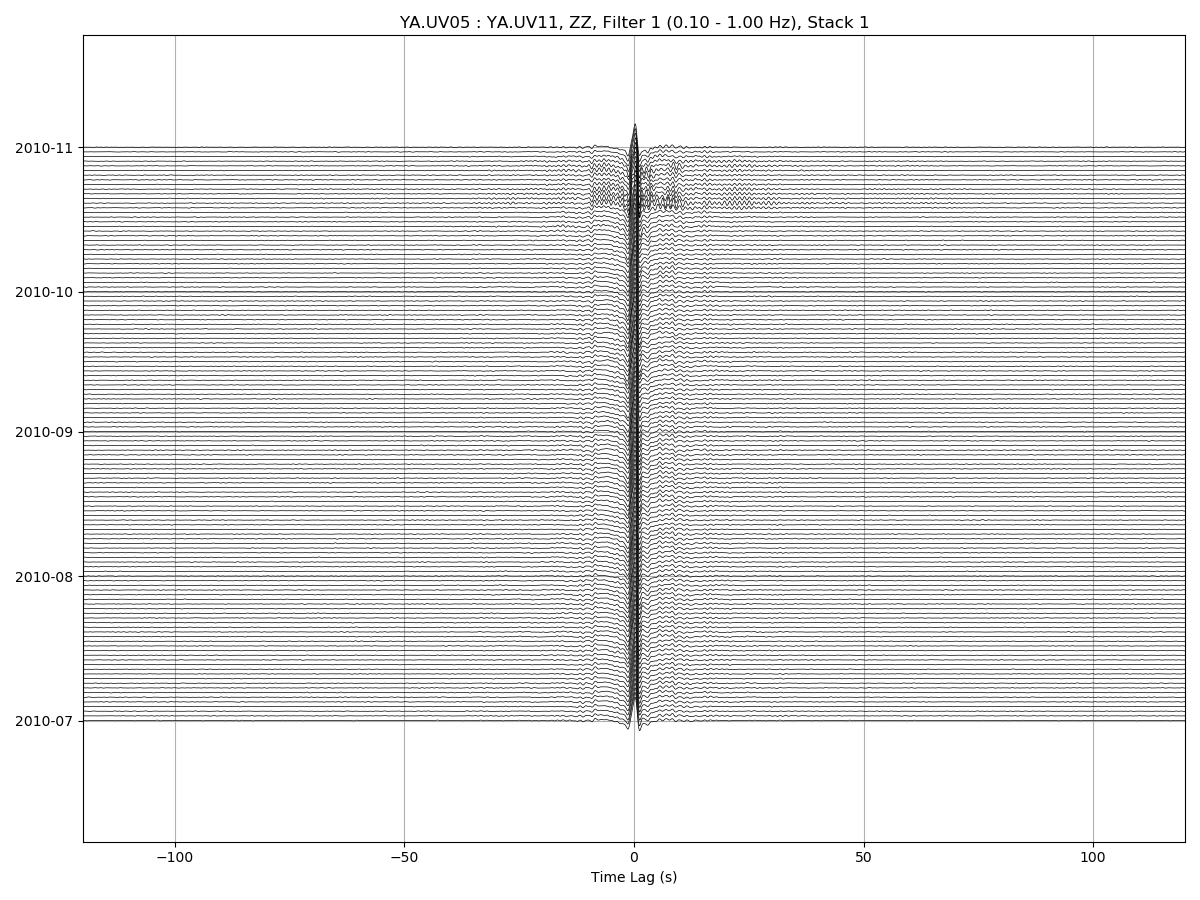
For zooming in the CCFs:
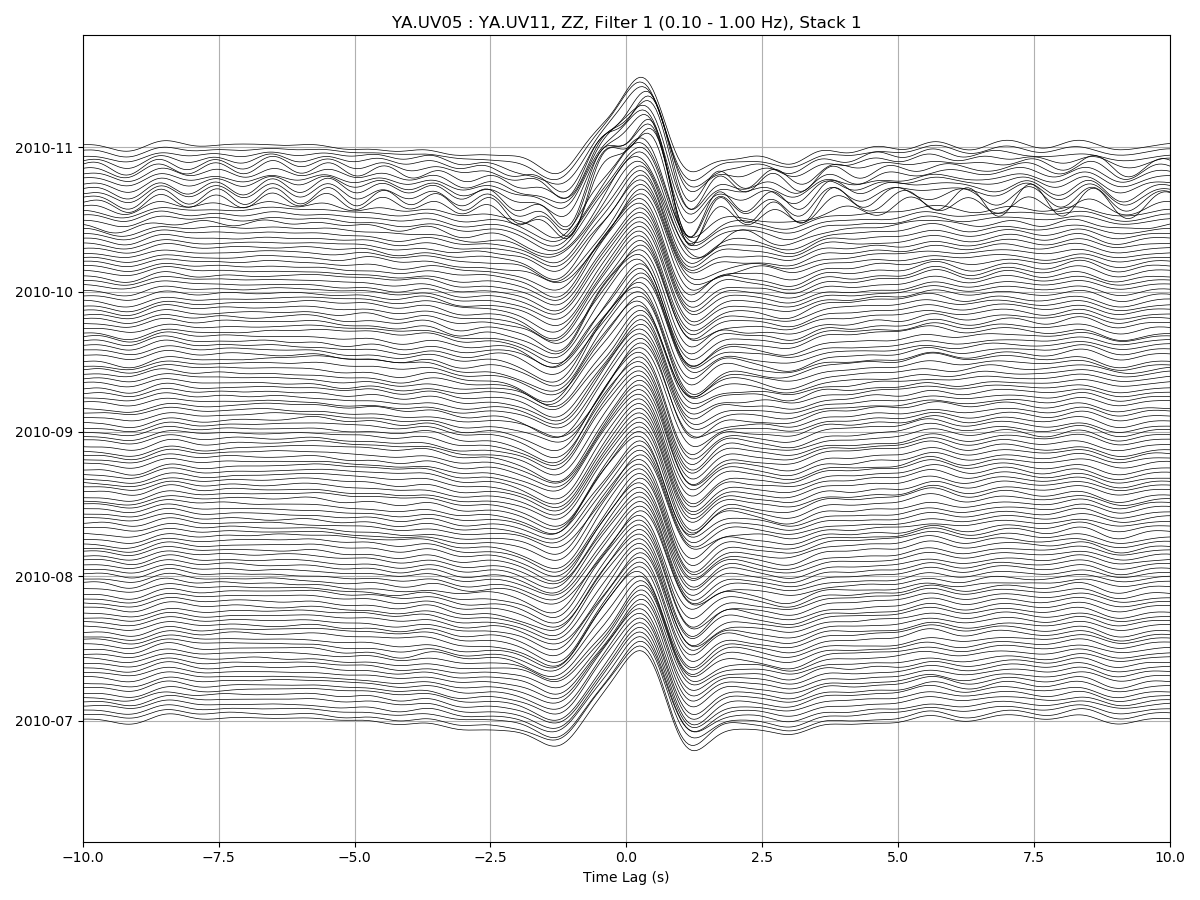
msnoise cc plot ccftime YA.UV05 YA.UV11 --xlim=-10,10 --ampli=15:
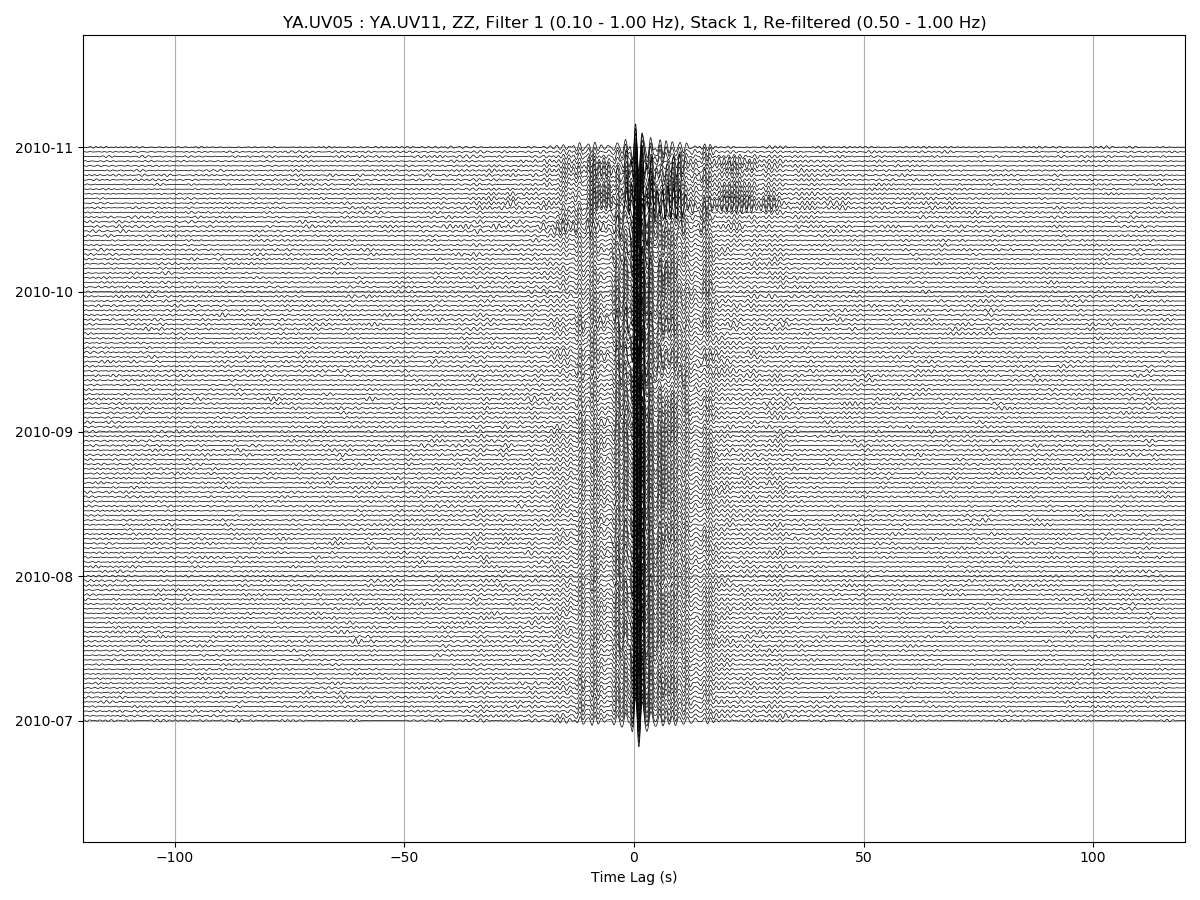
It is sometimes useful to refilter the CCFs on the fly:
msnoise cc plot ccftime YA.UV05 YA.UV11 -r 0.5:1.0:
→ Plot Interferograms
This plot shows the cross-correlation functions (CCF) vs time in a very similar
manner as on the ccftime plot above, but shows an image instead of wiggles.
The parameters allow to plot the daily or the mov-stacked CCF. Filters and
components are selectable too. Passing --refilter allows to bandpass filter
CCFs before plotting (new in 1.5).
msnoise cc plot interferogram --help
Usage: [OPTIONS] STA1 STA2 [EXTRA_ARGS]...
Plots the interferogram between sta1 and sta2 (parses the CCFs) STA1 and
STA2 must be provided with this format: NET.STA.LOC !
Options:
-f, --filterid INTEGER Filter ID
-c, --comp TEXT Components (ZZ, ZE, NZ, 1E,...). Defaults to ZZ
-m, --mov_stack INTEGER Mov Stack to read from disk. Defaults to 1.
-s, --show BOOLEAN Show interactively?
-o, --outfile TEXT Output filename (?=auto). Defaults to PNG format,
but can be anything matplotlib outputs, e.g. ?.pdf
will save to PDF with an automatic file naming.
-r, --refilter TEXT Refilter CCFs before plotting (e.g. 4:8 for
filtering CCFs between 4.0 and 8.0 Hz. This will
update the plot title.
--help Show this message and exit.
Example:
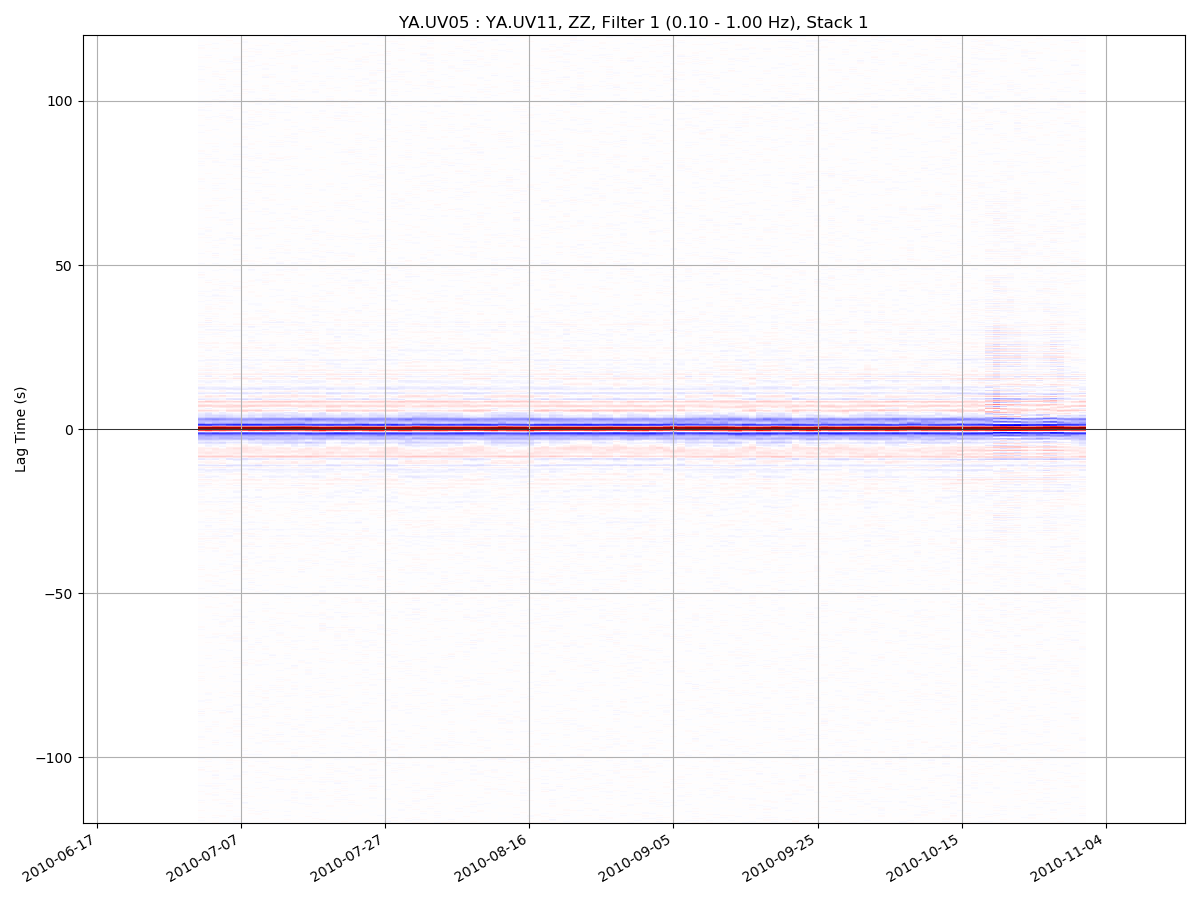
msnoise cc plot interferogram YA.UV06 YA.UV11 -m5 will plot the ZZ component
(default), filter 1 (default) and mov_stack 5:
→ Plot CCF’s spectrum vs Time
This plot shows the cross-correlation functions’ spectrum vs time. The
parameters allow to plot the daily or the mov-stacked CCF. Filters and
components are selectable too. The --ampli argument allows to increase the
vertical scale of the CCFs. Passing --refilter allows to bandpass filter
CCFs before computing the FFT and plotting. Passing --startdate and
--enddate parameters allows to specify which period of data should be
plotted. By default the plot uses dates determined in database.
msnoise cc plot spectime --help
Usage: [OPTIONS] STA1 STA2 [EXTRA_ARGS]...
Plots the ccf's spectrum vs time between sta1 and sta2 STA1 and STA2 must be
provided with this format: NET.STA.LOC !
Options:
-f, --filterid INTEGER Filter ID
-c, --comp TEXT Components (ZZ, ZE, NZ, 1E,...). Defaults to ZZ
-m, --mov_stack INTEGER Mov Stack to read from disk. Defaults to 1.
-a, --ampli FLOAT Amplification of the individual lines on the
vertical axis (default=1)
-s, --show BOOLEAN Show interactively?
-o, --outfile TEXT Output filename (?=auto). Defaults to PNG format,
but can be anything matplotlib outputs, e.g. ?.pdf
will save to PDF with an automatic file naming.
-r, --refilter TEXT Refilter CCFs before plotting (e.g. 4:8 for
filtering CCFs between 4.0 and 8.0 Hz. This will
update the plot title.
--help Show this message and exit.
Example:
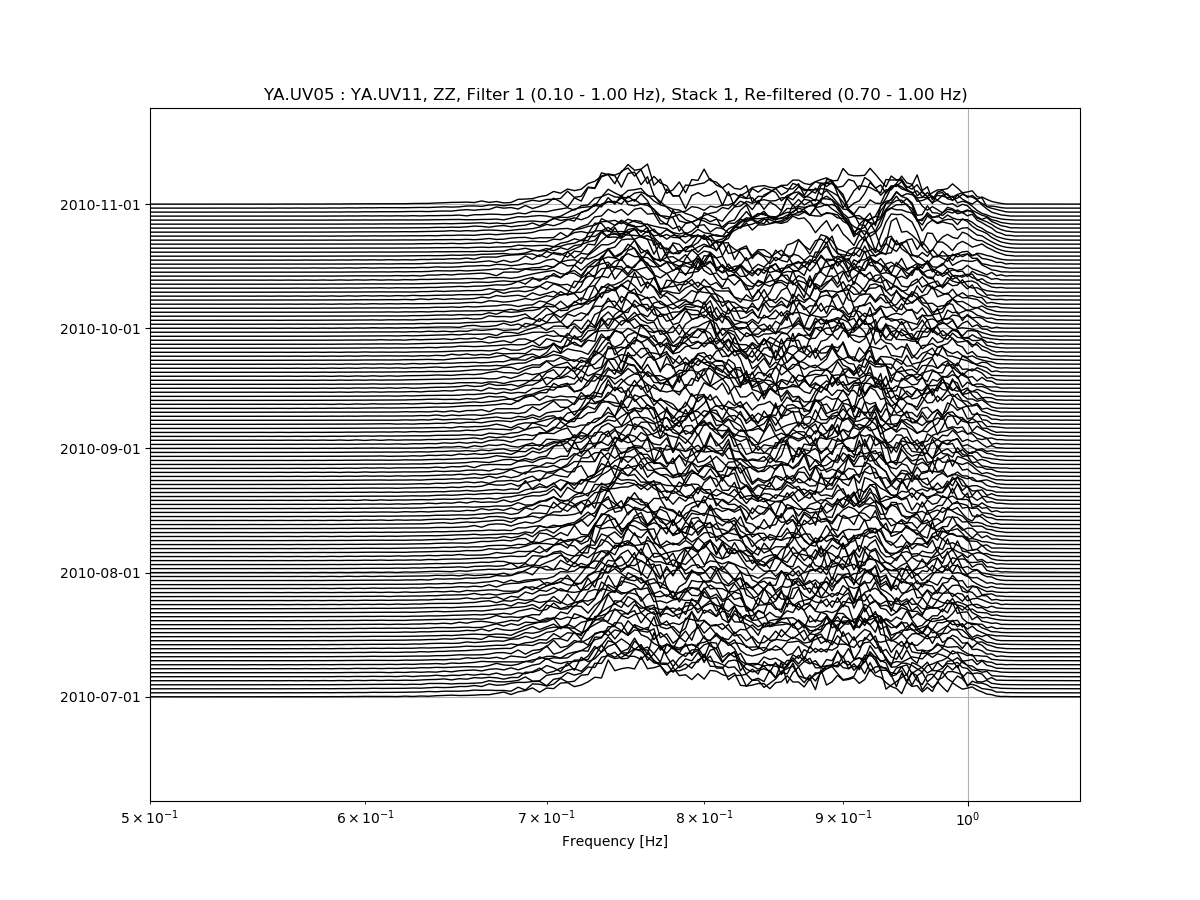
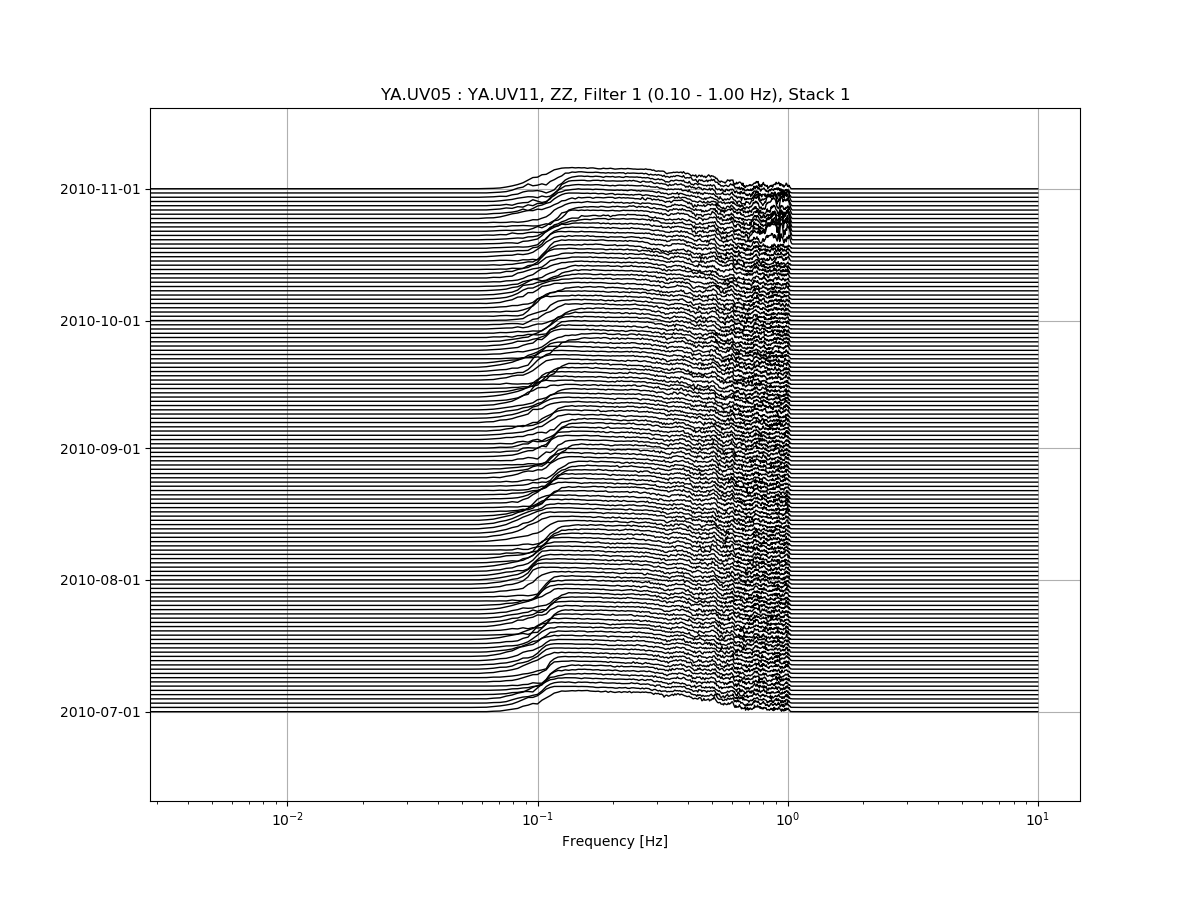
msnoise cc plot spectime YA.UV05 YA.UV11 will plot all defaults:
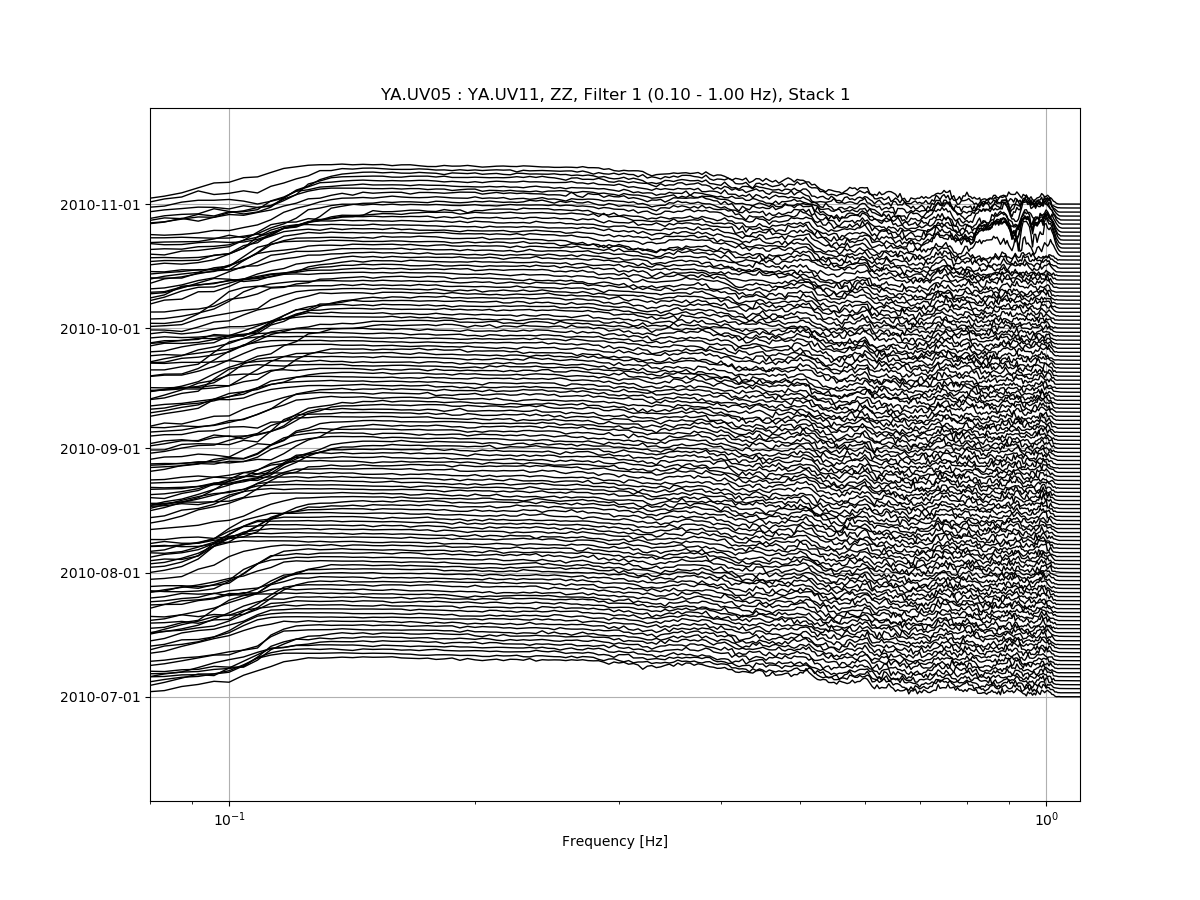
Zooming in the X-axis and playing with the amplitude:
msnoise cc plot spectime YA.UV05 YA.UV11 --xlim=0.08,1.1 --ampli=10:
And refiltering to enhance high frequency content:
msnoise cc plot spectime YA.UV05 YA.UV11 --xlim=0.5,1.1 --ampli=10 -r0.7:1.0: